For disease detection, every molecule counts.
Some genetic nuances are nearly undetectable in cell-free DNA
Every tissue releases cell-free DNA (cfDNA) into the bloodstream, a biomarker with limitless diagnostic potential. However, disease signals can be as low as 0.01%, often just a single molecule. To detect these, traditional methods amplify DNA millions of times, creating massive background noise that obscures the signal. As diagnostics shift from large, abundant chromosomal changes to sparse single-gene variations, conventional approaches hit a "noise wall" that fundamentally limits sensitivity.

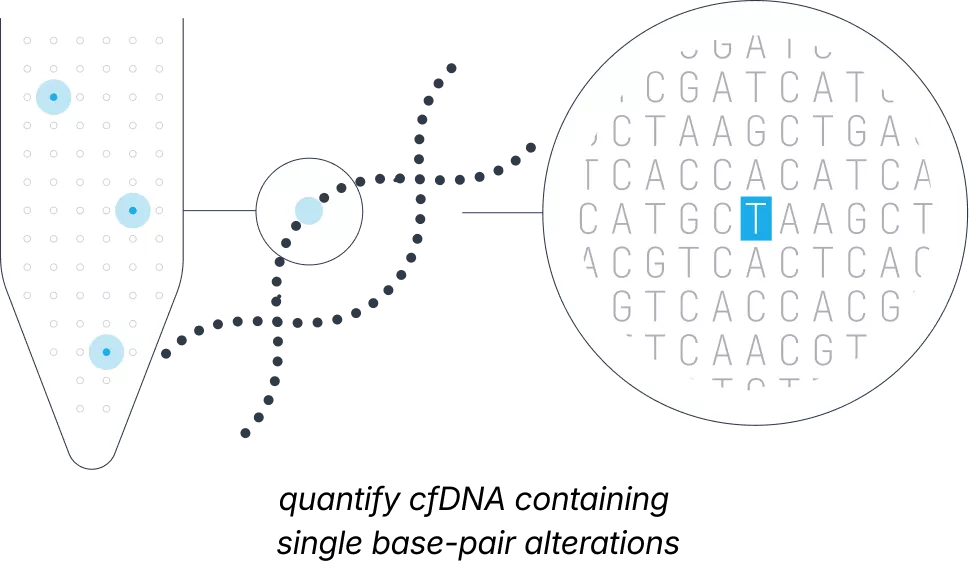
Traditional non-invasive prenatal screening focuses on assessing a fetus’ risk for larger chromosomal changes, such as trisomy 21. However, severe conditions like sickle cell disease result from tiny base-pair alterations within a single gene that are small and sparse compared to the vast maternal DNA in the background.
Our smNGS platform brings cfDNA diagnostics to true single-molecule resolution, quantifying these minute variations with molecular precision. By identifying these changes directly, UNITY Complete® provides personalized fetal risk assessment for single-gene conditions from a maternal blood draw as early as 9 weeks’ gestation, without requiring a partner sample.
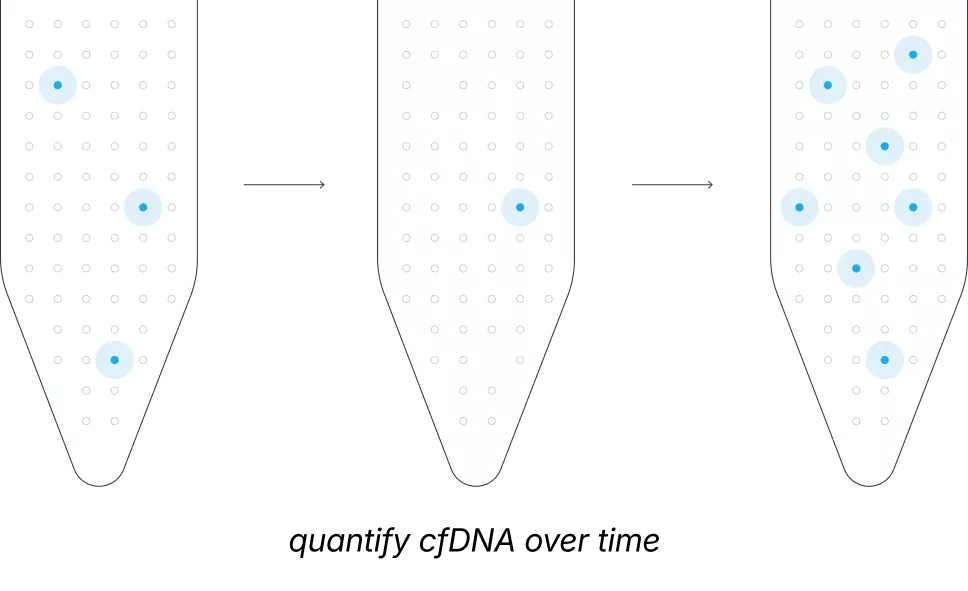
In cancer care, monitoring tumor evolution is critical, yet imaging often misses subtle molecular shifts. While liquid biopsy is a non-invasive alternative, it is hindered by a major biological hurdle: tumors often shed DNA into the bloodstream in extremely minute quantities. Most current assays fail to distinguish these trace signals from healthy genetic noise, limiting their use to high-burden or surgically accessible cases.
The Northstar suite uses smNGS to isolate true genomic signals with unprecedented precision. Northstar Select® overcomes low-shedding barriers to uncover over 50% more actionable tumor variants, while Northstar Response® tracks absolute tumor burden down to 0.01% tumor fraction to monitor treatment response in real-time.
Our smNGS Platform
We are transforming healthcare by redefining the limits of molecular diagnostics. Our proprietary single-molecule next-generation sequencing (smNGS) platform achieves the once-impossible: detecting and precisely quantifying genetic targets at the physical limit of detection— the single DNA molecule. Enabled by Quantitative Counting Templates™ (QCTs™), the platform quantifies disease-related DNA fragments with single base-pair resolution, providing absolute quantification where traditional methods only offer estimates.
Add
We add a specific number of traceable, synthetic DNA fragments (QCTs) into the patient’s blood sample, which contains an unknown number of target DNA fragments.

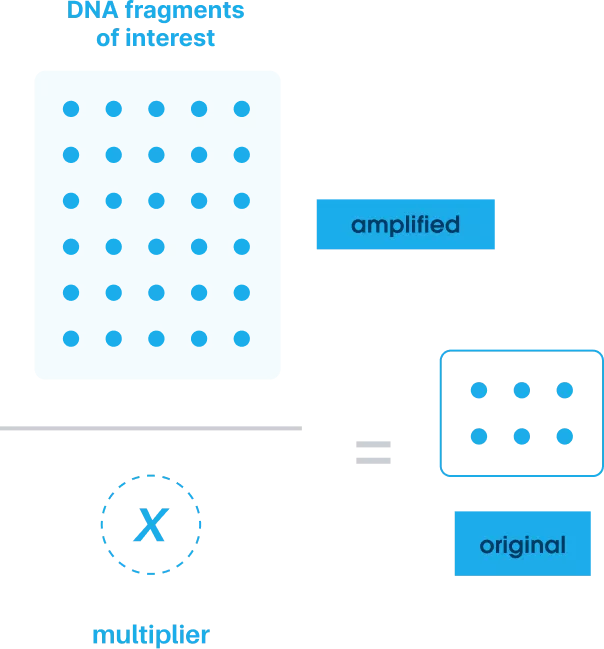
Amplify
Using PCR, the patient's DNA and the added QCTs are amplified at the same rate, ensuring the original ratio of molecules is preserved.
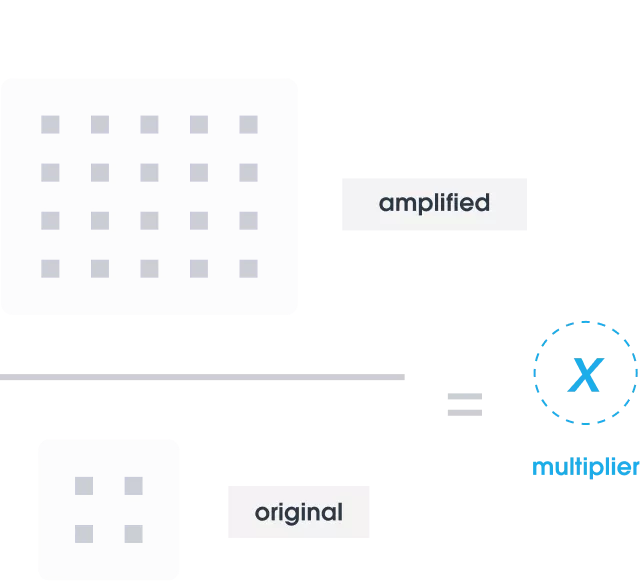
Count
We identify where noise occurs in the data and determine the amplification multiplier by dividing the number of QCTs found after sequencing by the number added initially.
Compute
Using AI-enhanced integrated workflows and proprietary bioinformatics, we apply this multiplier to determine the exact number of target DNA molecules present in the original sample.